Model that implements the linked cell algorithm. More...
#include <LinkedCells.h>
Public Member Functions | |
| LinkedCells (Force &force, double deltaT, std::array< double, 3 > domainSize, double rCutOff, FileHandler::outputFormat outputFormat, BoundarySet boundaryConditions, bool gravityOn, std::array< double, 3 > g={}, MembraneParameters membraneParameters=MembraneParameters{}, enumsStructs::ParallelizationStrategy parallelizationStrategy=enumsStructs::ParallelizationStrategy::none) | |
| Contruct a new Linked Cells model. More... | |
| void | step (int iteration) override |
| Perform one time step in the linked cells model. More... | |
| void | updateForcesOptimized () |
| Implements the optimization we presented as our second idea. At the moment this is dead code, because we did not have time yet to make it compatible with our current program structure and only integrated it once when doing the time measurements. More... | |
| void | initializeForces () override |
| Calculate forces at the beginning of the simulation that the old force is not 0. More... | |
 Public Member Functions inherited from Model Public Member Functions inherited from Model | |
| virtual | ~Model ()=default |
| Virtual default constructor to guarantee appropriate memory clean up. More... | |
| void | plot (int iteration, std::string &baseName) |
| Write current state of the model to a file. More... | |
| void | addCuboid (const std::array< double, 3 > &position, unsigned N1, unsigned N2, unsigned N3, double h, double mass, const std::array< double, 3 > &initVelocity, int dimensions, double brownianMotionAverageVelocity, double epsilon=5, double sigma=1, bool fixed=false) |
| Add a cuboid structure to this model. More... | |
| void | addDisc (const std::array< double, 3 > ¢er, const std::array< double, 3 > &initVelocity, int N, double h, double mass, int dimensions, double brownianMotionAverageVelocity, double epsilon=5, double sigma=1, bool fixed=false) |
| Add a 2D disc structure to this model. More... | |
| void | addSphere (const std::array< double, 3 > ¢er, const std::array< double, 3 > &initVelocity, int N, double h, double mass, int dimensions, double brownianMotionAverageVelocity, double epsilon=5, double sigma=1, bool fixed=false) |
| Add a 3D sphere structure to this model. More... | |
| void | addParticle (Particle &p) |
| Add a single particle to this model. More... | |
| void | addViaFile (std::string &filepath, FileHandler::inputFormat inputFormat) |
| Add new particles / particle structures to the model via a file. More... | |
| void | saveState () |
| Export the current state of all molecules to a txt file for using them in a new simulation. More... | |
| void | updateForces () const |
| Helper method to calculate the force between all particles. More... | |
| ParticleContainer & | getParticles () const |
| Get the Particles of this model. More... | |
Private Member Functions | |
| void | processBoundaryForces () |
| Apply forces to all particles in boundary cells according to the specified boundary conditions. More... | |
| void | processHaloCells () |
| Process all particles which have left the domain at the end of a simulation step according to the specified boundary conditions. More... | |
| void | pullMarkedParticles () |
| Apply to all particles that are marked a constant force. More... | |
| void | updateForcesMembrane () |
| Calculate forces between particles in a membrane structure. More... | |
Private Attributes | |
| bool | membraneSetting |
| bool | pull |
| int | pullingActiveUntil |
| std::array< double, 3 > | pullingForce {} |
| std::unique_ptr< Force > | forceBetweenDiagonalNeighborsInMembrane |
| std::unique_ptr< Force > | forceBetweenDirectNeighborsInMembrane |
| enumsStructs::ParallelizationStrategy | parallelizationStrategy |
| LinkedCellsContainer | particles |
| std::vector< std::pair< Side, enumsStructs::BoundaryCondition > > | boundarySettings |
Additional Inherited Members | |
 Protected Member Functions inherited from Model Protected Member Functions inherited from Model | |
| Model (ParticleContainer &particles, Force &force, double deltaT, FileHandler::outputFormat outputFormat, bool gravityOn, std::array< double, 3 > g={}) | |
| Constructor for this model. Cannot be called from any other class but classes that extend this class, because this class is a base class and should not be instantiated. More... | |
| void | updatePositions () const |
| Helper method to calculate the position of all particles. More... | |
| void | updateVelocities () const |
| Helper method to calculate the velocity of all particles. More... | |
| void | applyGravity () |
| Add a gravitational force of g * m along the y-axis to each particle inside the simulation domain. More... | |
 Protected Attributes inherited from Model Protected Attributes inherited from Model | |
| ParticleContainer & | particles |
| Force & | force |
| double | deltaT |
| bool | gravityOn |
| std::array< double, 3 > | g |
Detailed Description
Model that implements the linked cell algorithm.
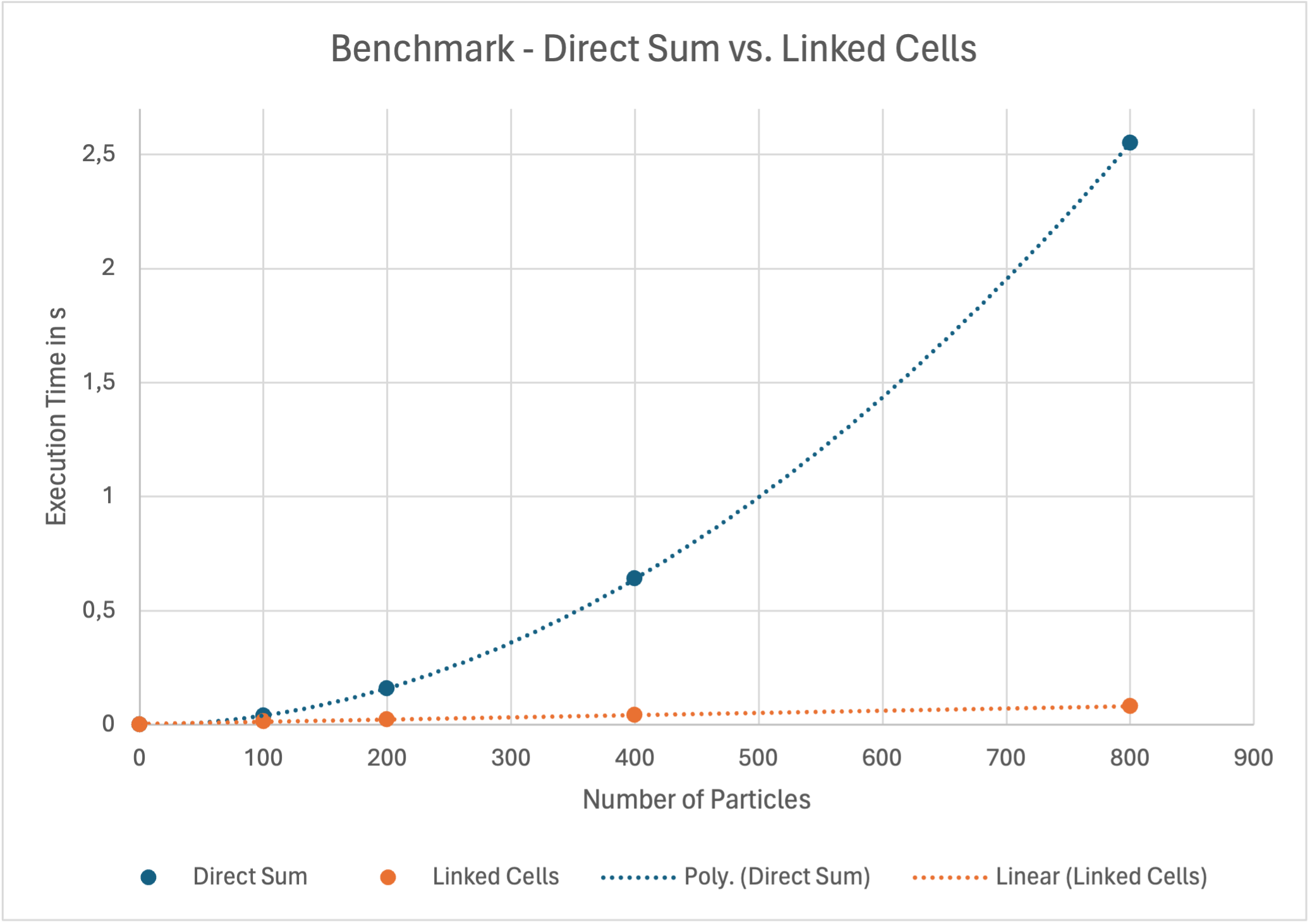
The linked-cells algorithm is significantly faster than the direct sum algorithm. It reduces the time complexity from O(n²) to O(n). The following plot shows some benchmarks.
Constructor & Destructor Documentation
◆ LinkedCells()
| LinkedCells::LinkedCells | ( | Force & | force, |
| double | deltaT, | ||
| std::array< double, 3 > | domainSize, | ||
| double | rCutOff, | ||
| FileHandler::outputFormat | outputFormat, | ||
| BoundarySet | boundaryConditions, | ||
| bool | gravityOn, | ||
| std::array< double, 3 > | g = {}, |
||
| MembraneParameters | membraneParameters = MembraneParameters{}, |
||
| enumsStructs::ParallelizationStrategy | parallelizationStrategy = enumsStructs::ParallelizationStrategy::none |
||
| ) |
Contruct a new Linked Cells model.
- Parameters
-
force Force to use. deltaT Discretisation step. domainSize Domain size. rCutOff Cutoff radius. outputFormat Output format. boundaryConditions Boundary conditions. gravityOn Toggle gravity on or off. g Gravitational factor. membraneParameters Contains specification if a membrane is simulated parallelizationStrategy Strategy to use for parallelization.
Member Function Documentation
◆ initializeForces()
|
overridevirtual |
Calculate forces at the beginning of the simulation that the old force is not 0.
Implements Model.
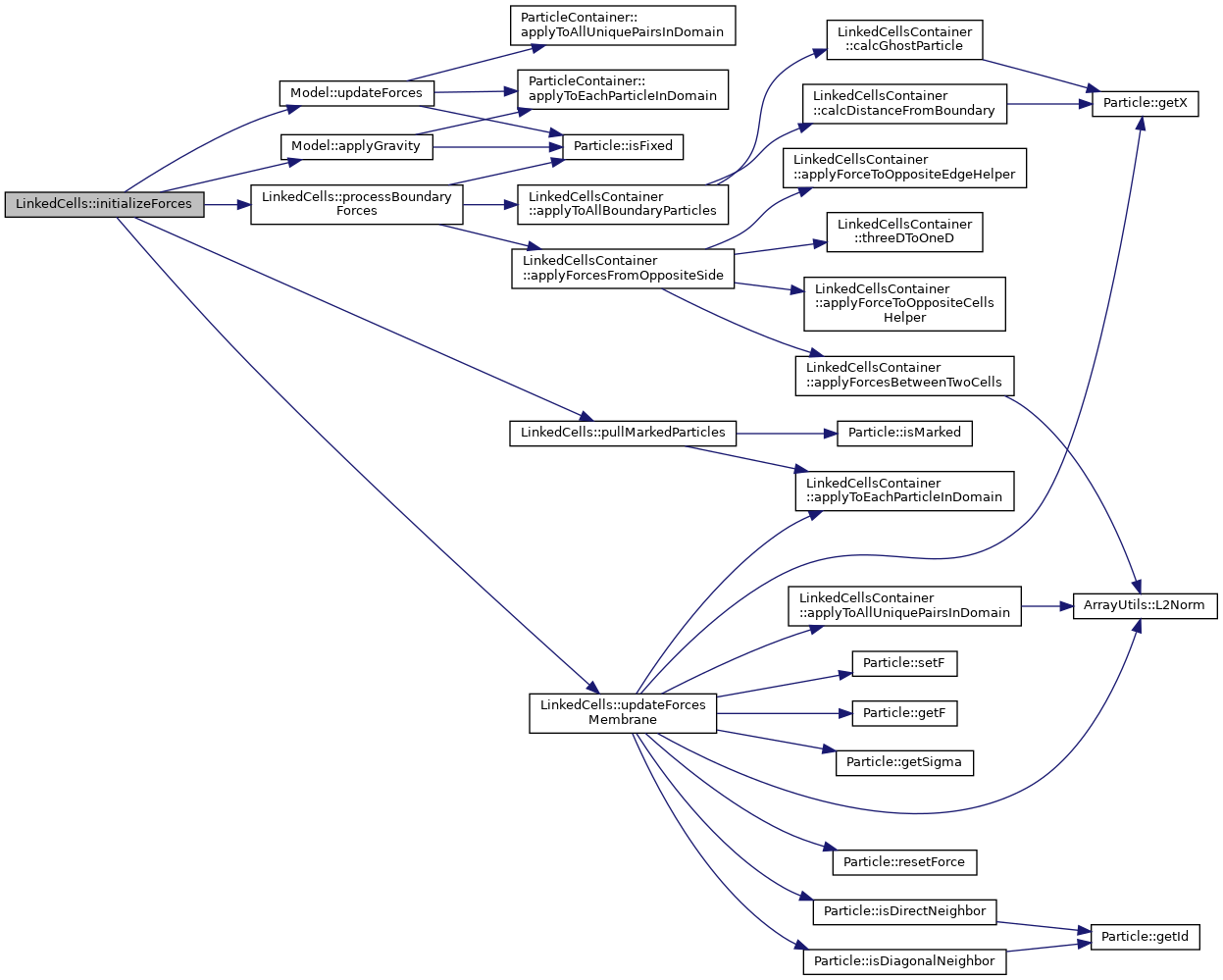
◆ processBoundaryForces()
|
private |
Apply forces to all particles in boundary cells according to the specified boundary conditions.
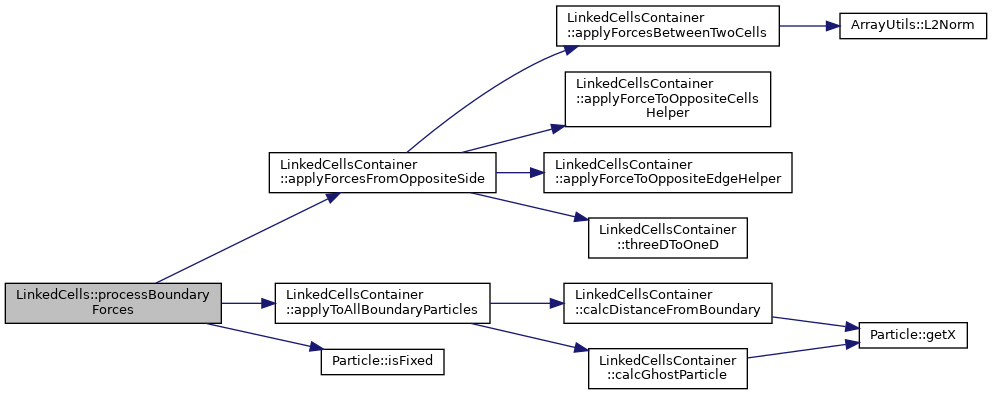
◆ processHaloCells()
|
private |
Process all particles which have left the domain at the end of a simulation step according to the specified boundary conditions.
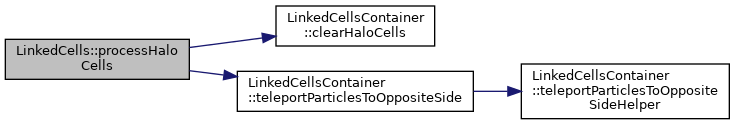
◆ pullMarkedParticles()
|
private |
Apply to all particles that are marked a constant force.
◆ step()
|
overridevirtual |
Perform one time step in the linked cells model.
- Parameters
-
iteration Current iteration the simulator is in.
Implements Model.
◆ updateForcesMembrane()
|
private |
Calculate forces between particles in a membrane structure.
◆ updateForcesOptimized()
| void LinkedCells::updateForcesOptimized | ( | ) |
Implements the optimization we presented as our second idea. At the moment this is dead code, because we did not have time yet to make it compatible with our current program structure and only integrated it once when doing the time measurements.
Member Data Documentation
◆ boundarySettings
|
private |
Definition of the boundary condition for each side.
◆ forceBetweenDiagonalNeighborsInMembrane
|
private |
◆ forceBetweenDirectNeighborsInMembrane
|
private |
◆ membraneSetting
|
private |
◆ parallelizationStrategy
|
private |
◆ particles
|
private |
This model uses the LinkedCellsContainer to store its particles
◆ pull
|
private |
◆ pullingActiveUntil
|
private |
◆ pullingForce
|
private |
The documentation for this class was generated from the following files:
- /home/runner/work/MolSim/MolSim/src/models/linkedCells/LinkedCells.h
- /home/runner/work/MolSim/MolSim/src/models/linkedCells/LinkedCells.cpp